Introduction
Networks are abundant in many areas of biology. These networks often entail non-trivial topological features and patterns critical to understanding interactions within the natural system. However, networks observed in real-world are typically noisy. The presence of high levels of noise can hamper discovery of structures and dynamics present in the network.
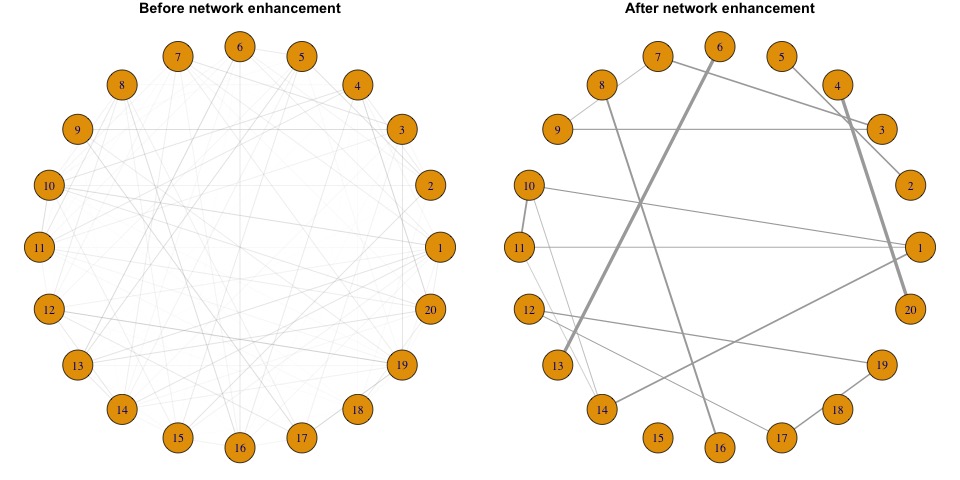
Wang et al. propose Network Enhancement (NE), a novel method for improving the signal-to-noise ratio of a symmetric networks and thereby facilitating the downstream network analysis. NE leverages the transitive edges of a network by exploiting local structures to strengthen the signal within clusters and weaken the signal between clusters. At the same time NE also alleviates the corrupted links in the network by imposing a normalization that removes weak edges by enforcing sparsity. NE is supported by theoretical justifications for its convergence and performance in improving community detection outcomes.
The method provides theoretical guarantees as well as excellent empirical performance on many biological problems. The approach can be incorporated into any weighted network analysis pipeline and can lead to improved downstream analysis.
We translated the matlab code for network enhancement into a R package named $neten$
The network enhancement repository
The network enhancement project website
Installation
You can install it from GitHub with devtools:
install_github("microbma/neten")
Or you can download the binary package for this package from GitHub repository:
https://github.com/microbma/neten
Useage
The $Network_Enhancement$ command require to input a correlation matrix, either directed or undirected.
mat1 <- cor(matrix(rnorm(400),nrow = 20))
mat2 <- Network_Enhancement(mat1)
mat2 is the enhanced matrix. From this matrix we can infer a network with higher confidence.